Animal Coronaviruses and the Susceptibility to COVID-19 in Humans
Coronaviruses (CoV) were first discovered in the late 1930s and were mainly known to cause diarrhea in young animals and mild respiratory illness in human and animal hosts. The outbreak of SARS-CoV in 2003 and MERS-CoV in 2012 caused worldwide epidemics with hundreds of deaths. The evolution of coronaviruses responsible for severe respiratory distress syndrome showed the threatening and devastating potential of new CoV strains.
Currently, we are on the heels of SARS and MERS, in the middle of a third wave of severe respiratory distress syndrome. SARS-CoV2, also known as COVID-19, started a worldwide pandemic in December 2019, with hundreds of thousands of deaths, setting hundreds of countries into states of emergency (Ye et al., 2020). We need immediate treatment or prophylactic strategies to control the spread of the disease. The understanding of animal-human transmission and cross immunity between human and animal strains is of utmost importance for the development of vaccines.
Coronaviruses are part of the family Coronaviridae, which are characterised as enveloped and positive sensed, single-stranded RNA viruses. They originate from bats, rodents or birds but have an extremely broad range of host species spread globally. A high mutation and recombination rate as well as the largest genome of all RNA-viruses enables them to evolve quickly and adapt regularly to new hosts and environments (Bolles, Donaldson & Baric, 2011).
Coronaviruses are separated into four main groups Alpha, Beta, Gamma and Delta. Human severe acute respiratory syndromes SARS, MERS and SARS-CoV2 all originated from the Beta genus, which is divided further into subgenera A, B, C and D. MERS is classified in group Beta C. Group Beta B includes SARS-CoV and SARS-CoV2 (COVID-19). Group Beta A encompasses a large number of different species (Luk, Li, Fung, Lau & Woo, 2019). Most important for this study are equine, bovine and canine coronaviruses (ECoV, BCoV and CCoV) which cause mild respiratory and/or gastrointestinal symptoms depending on the strain (Decaro et al., 2013; Jenkinson et al., 2001; Miszczak et al., 2014; Mitchell et al., 2017).
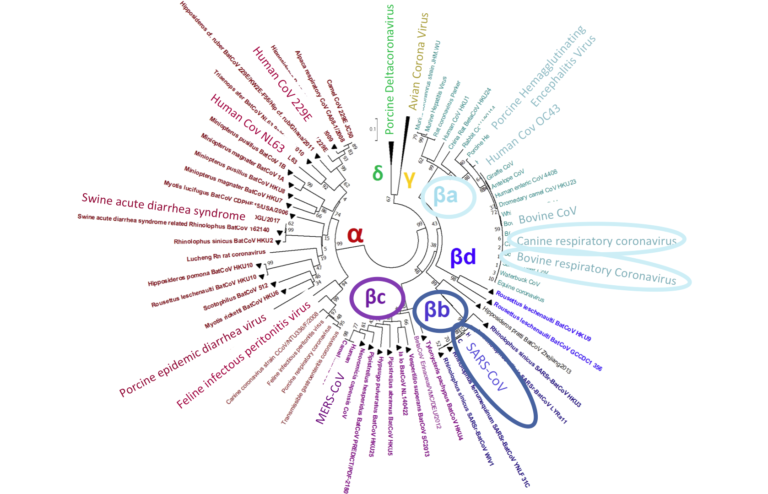 A phylogenetic tree illustrating the diversity of coronaviruses and their subgroups (adapted from Luk et al., 2019) (click to enlarge)
A phylogenetic tree illustrating the diversity of coronaviruses and their subgroups (adapted from Luk et al., 2019) (click to enlarge)
All three animal coronaviruses circulate in Europe. BCoV antigen was detected by PCR in 47% of examined Swiss cattle (Meylan, 2020, unpublished) as was ECoV at 1.2-2% prevalence in French and Irish horses (Miszczak et al., 2014; Nemoto, Schofield & Cullinane, 2019). In a European study, a sero-prevalence of 47% was found in dogs (Mitchell et al., 2017). ECoV, BCoV and CCoV are very closely related to OC43 a mild respiratory corona disease in humans. Phylogenetic analysis showed that OC43 evolved relatively recently, around 1890, from BCoV (Vijgen et al., 2005).
Cross-reactivity between phylogenetic closely related species does exist. It has been proven between CoVs of the same subgenus (Gerdts & Zakhartchouk, 2017; Goede et al., 2015; Jenkinson et al., 2001) and an interspecies protective effect could even be proven, provided it was within the same genus (Han, Cheon, Zhang & Saif, 2006; Woods & Pedersen, 1979).
A comparative study published in April 2020 compared nucleocapsid epitopes of different corona strains to COVID-19. Nucleocapsids are the most common protein group in coronaviruses. They are involved in replication and transcription processes and the packing of the viral genome. Additionally, they hinder the reproductive cycle of the host cell. More importantly, nucleocapsid epitopes (NCEs) prove to provoke strong immune reactions, and the NCE amino acid sequence is normally well conserved in the genome. This makes NCEs the perfect candidate for science in vaccine and diagnostic development. The study’s focus was NCEs known to provoke a strong T cell and antibody response in the host. Five NCE domains were detected to be very similar between COVID-19, bovine- and canine CoVs, despite a low similarity on the whole NC-sequence (Tilocca et al., 2020).
As BCoV, CCoV and ECoV are in the same genus as COVID-19 and have been present in Switzerland for longer time periods, we hypothesise that sub-clinical exposure between humans and animals could have a protective effect for COVID-19 through contact with animals.
Further reading
Decaro, N. & Lorusso, A. (2020). Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses, in: Veterinary Microbiology, 244, 108693.
Padron-Regalado, E. (2020). Vaccines for SARS-CoV-2: Lessons from Other Coronavirus Strains, in: Infectious Diseases and Therapy.
Sheikh, A., Al-Taher, A., Al-Nazawi, M., Al-Mubarak, A. I. & Kandeel, M. (2020). Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design, in: Journal of Virological Methods, 277, 113806.
Shi, J., Wen, Z., Zhong, G., Yang, H., Wang, C., Huang, B., […] Bu, Z. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2, in: Science, 368, 6949, 1016-1020.
References
Bolles, M., Donaldson, E. & Baric, R. (2011). SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission, in: Current Opinion in Virology, 1, pp. 624–634.
Decaro, N., Cordonnier, N., Demeter, Z., Egberink, H., Elia, G., Grellet, A., […] Buonavoglia, C. (2013). European surveillance for pantropic canine coronavirus, in: Journal of Clinical Microbiology, 51(1), 83–88.
Gerdts, V. & Zakhartchouk, A. (2017). Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses, in: Veterinary Microbiology, 206, 45–51.
Goede, D., Murtaugh, M. P., Nerem, J., Yeske, P., Rossow, K. & Morrison, R. (2015). Previous infection of sows with a “mild” strain of porcine epidemic diarrhea virus confers protection against infection with a “severe” strain, in: Veterinary Microbiology, 176(1–2), 161–164.
Han, M. G., Cheon, D.-S., Zhang, X. & Saif, L. J. (2006). Cross-Protection against a Human Enteric Coronavirus and a Virulent Bovine Enteric Coronavirus in Gnotobiotic Calves, in: Journal of Virology, 80(24), 12350–12356.
Jenkinson, C. P., Hanson, R., Cray, K., Wiedrich, C., Knowler, W. C., Bogardus, C. & Baier, L. (2001). Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection, in: Archives of Virology, 146(12), 2401–2419.
Luk, H. K. H., Li, X., Fung, J., Lau, S. K. P. & Woo, P. C. Y. (2019). Molecular epidemiology, evolution and phylogeny of SARS coronavirus, in: Infection, genetics and evolution, 71, 21-30.
Miszczak, F., Tesson, V., Kin, N., Dina, J., Balasuriya, U. B. R., Pronost, S. & Vabret, A. (2014). First detection of equine coronavirus (ECoV) in Europe, in: Veterinary Microbiology, 171(1–2), 206–209.
Mitchell, J. A., Cardwell, J. M., Leach, H., Walker, C. A., Le Poder, S., Decaro, N., […] Brownlie, J. (2017). European surveillance of emerging pathogens associated with canine infectious respiratory disease, in: Veterinary Microbiology, 212, 31–38.
Nemoto, M., Schofield, W. & Cullinane, A. (2019). The first detection of equine coronavirus in adult horses and foals in Ireland, in: Viruses, 11(10).
Tilocca, B., Soggiu, A., Sanguinetti, M., Musella, V., Britti, D., Bonizzi, L., […] Roncada, P. (2020). Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses, in: Microbes and Infection, 7.
Vijgen, L., Keyaerts, E., Moës, E., Thoelen, I., Wollants, E., Lemey, P., […] Van Ranst, M. (2005). Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event, in: Journal of Virology, 79(3), 1595–1604.
Woods, R. D. & Pedersen, N. C. (1979). Cross-protection studies between feline infectious peritonitis and porcine transmissible gastroenteritis viruses, in: Veterinary Microbiology, 4(1), 11–16.
Ye, Z.-W., Yuan, S., Yuen, K.-S., Fung, S.-Y., Chan, C.-P. & Jin, D.-Y. (2020). Zoonotic origins of human coronaviruses, in: International Journal of Biological Sciences, (10), 1686–1697.
Share this
One Health: Connecting Humans, Animals and the Environment

One Health: Connecting Humans, Animals and the Environment


Reach your personal and professional goals
Unlock access to hundreds of expert online courses and degrees from top universities and educators to gain accredited qualifications and professional CV-building certificates.
Join over 18 million learners to launch, switch or build upon your career, all at your own pace, across a wide range of topic areas.
Register to receive updates
-
Create an account to receive our newsletter, course recommendations and promotions.
Register for free







