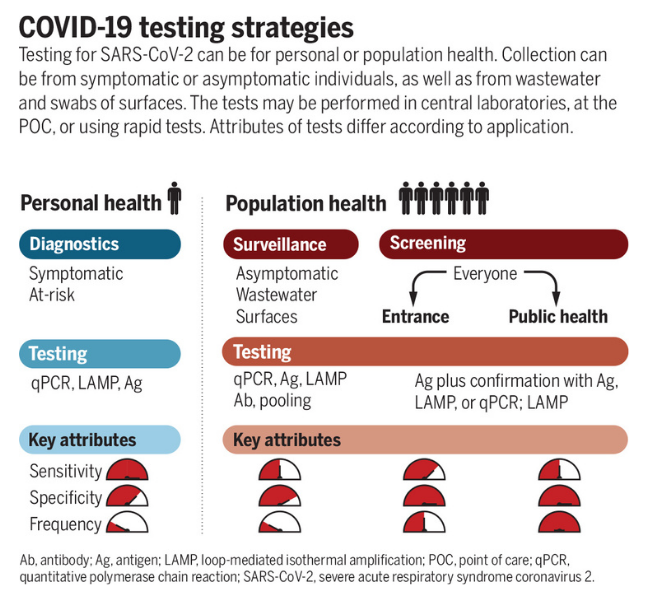
Home / Healthcare & Medicine / Biology & Biotechnology / From Swab to Server: Testing, Sequencing, and Sharing During a Pandemic / Sample types for testing
This article is from the free online
From Swab to Server: Testing, Sequencing, and Sharing During a Pandemic

Reach your personal and professional goals
Unlock access to hundreds of expert online courses and degrees from top universities and educators to gain accredited qualifications and professional CV-building certificates.
Join over 18 million learners to launch, switch or build upon your career, all at your own pace, across a wide range of topic areas.