Home / Science, Engineering & Maths / Biology & Biotechnology / The Biology of Bugs, Brains, and Beasts / Biology toolbox: Enzyme-substrate interactions and inhibition

Reach your personal and professional goals
Unlock access to hundreds of expert online courses and degrees from top universities and educators to gain accredited qualifications and professional CV-building certificates.
Join over 18 million learners to launch, switch or build upon your career, all at your own pace, across a wide range of topic areas.


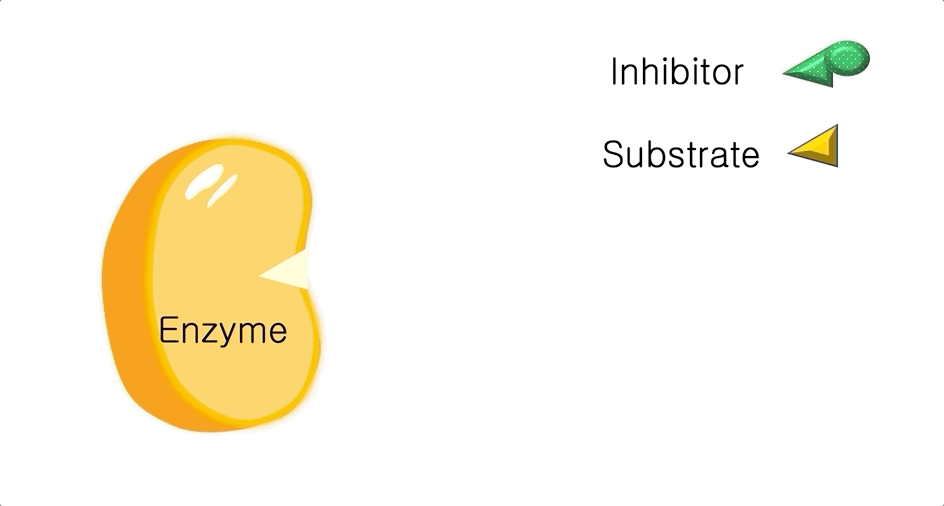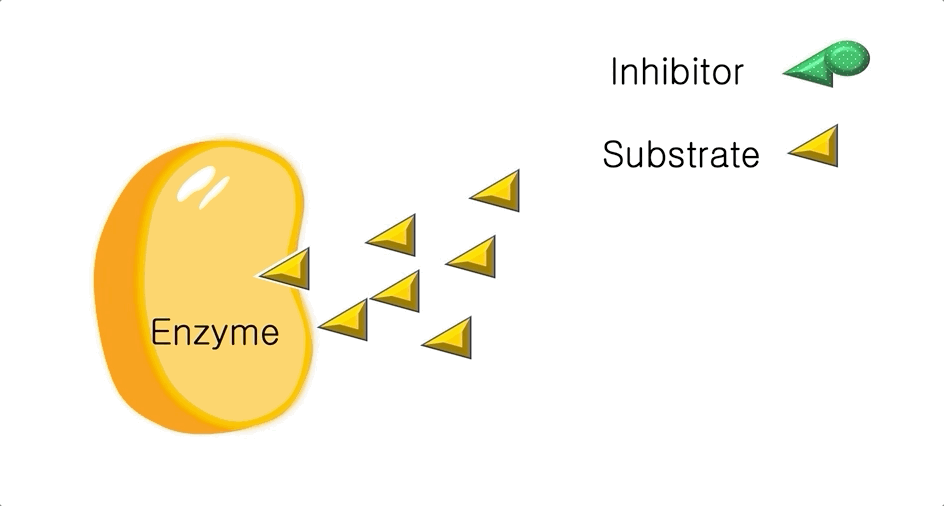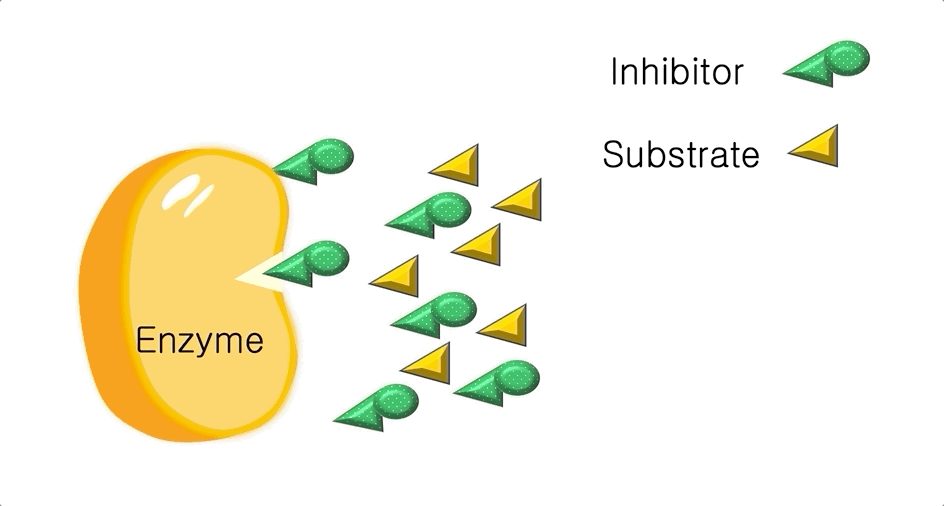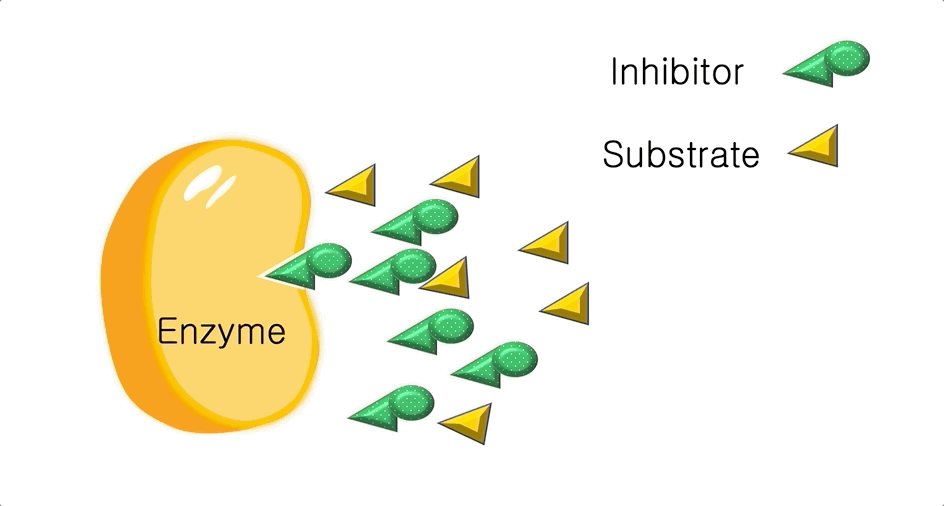
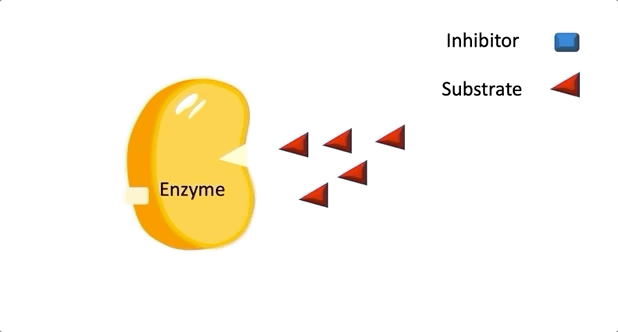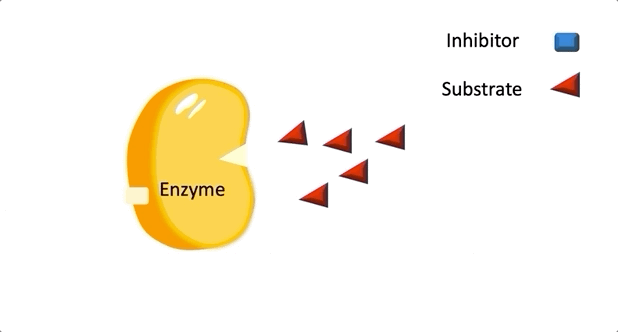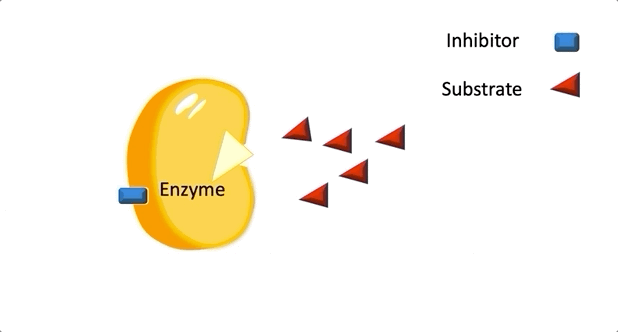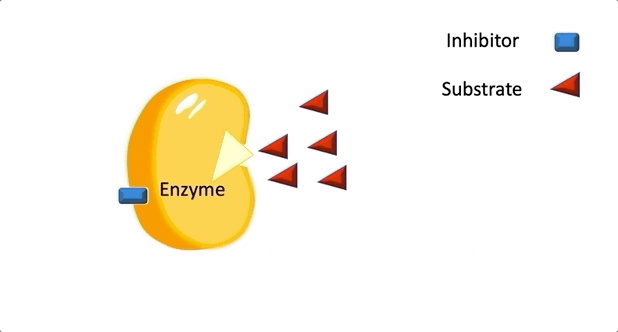
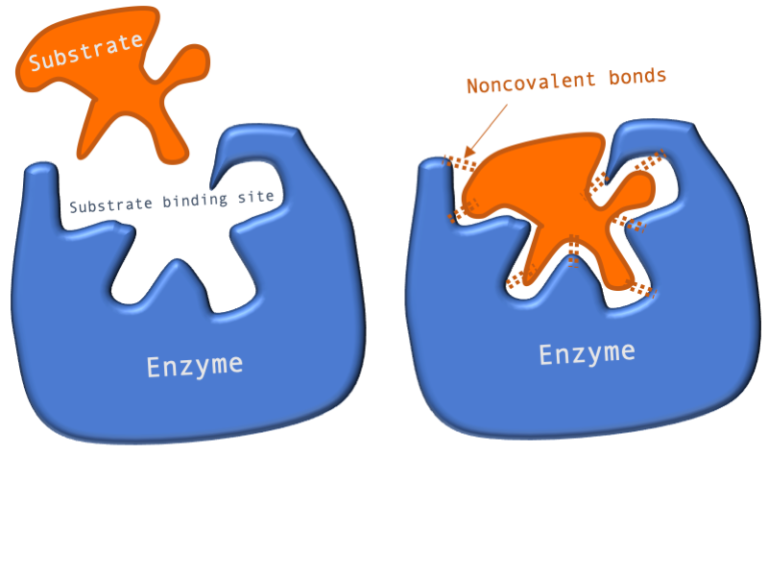 Figure 1: A diagram showing the Lock and Key model of enzyme-substrate interaction.
Figure 1: A diagram showing the Lock and Key model of enzyme-substrate interaction.
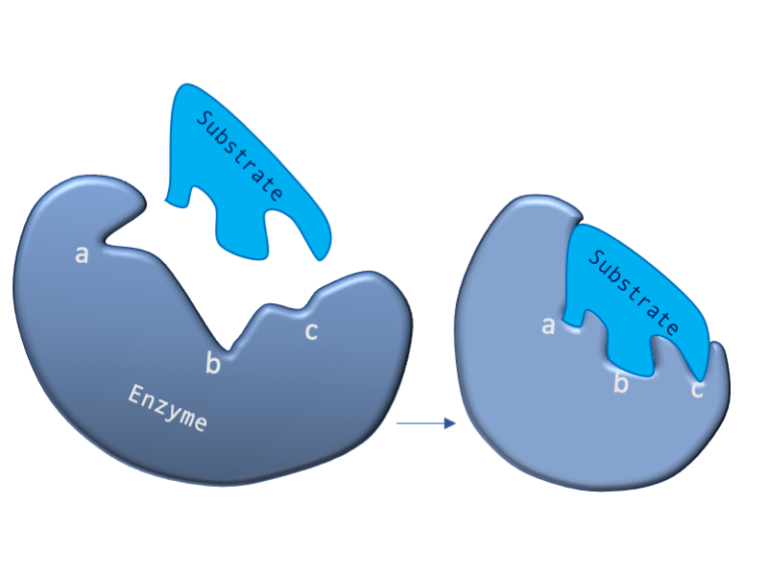 Figure 2: A diagram showing the Induced Fit model of enzyme substrate interaction
Figure 2: A diagram showing the Induced Fit model of enzyme substrate interaction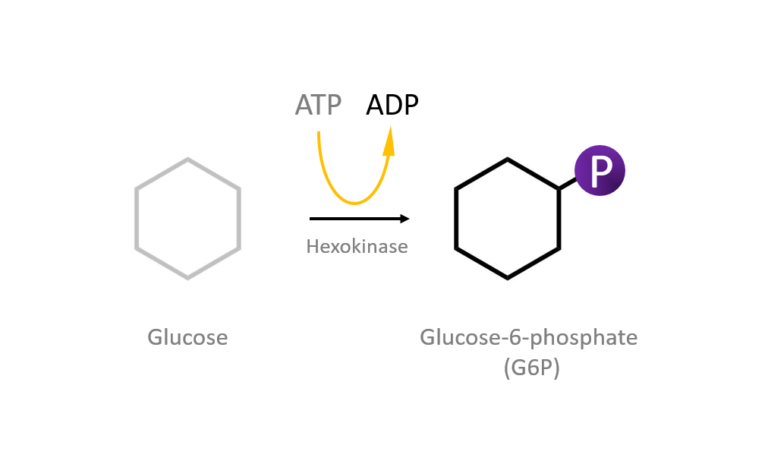 Figure 3: Hexokinase converts glucose to glucose 6-phosphate during glycolysis Note: G6P is very structurally similar to glucose (G6P= glucose with an added phosphate group).
Figure 3: Hexokinase converts glucose to glucose 6-phosphate during glycolysis Note: G6P is very structurally similar to glucose (G6P= glucose with an added phosphate group).