Competency-Based Design and Competency Frameworks
Competency mapping is a process of identifying key competencies needed to successfully carry out a specific job or set of tasks.
Several competency frameworks (CF) for genomics and bioinformatics exist based on professional roles from researchers to healthcare professionals. In this step we will discuss how CFs can be applied to designing courses, although they can also be used to revise and map existing courses to determine gaps in competencies which in turn can also lead to the design of new modules or courses.
Competency framework (CF) for bioinformatics
The International Society of Computational Biology (ISCB) have developed a framework which defines the core competencies required by professionals working in fields related to bioinformatics and computational biology. You can find the latest published version at the linked page, though this competency framework is still being developed and is evolving. The latest version can be found here.
The ISCB competency framework specifies 16 competencies (labelled A to P), capturing the scientific, technical and professional competencies required by individuals working in different roles that have a significant requirement for bioinformatics (please see the table below). Each of the competencies A-P can be further unravelled to unveil more detailed competencies within the general one. Altogether, this set of competencies defines Knowledge, Skills and Attitudes (KSA) desirable for a specific role together with a level of KSA required for each specific competency (more on this later).
The widely defined roles or personas captured in the framework include bioinformatics users (physician, lab technician, ethicist and bio curator), bioinformatics scientists (life science researcher, educator, academic researcher and core facility scientists) and bioinformatics engineer (bioinformatician in research support role or software developer). An expected level of competence (as in Bloom’s taxonomy, see Step 1.17) is associated to each role for each competency A-P. Here is the table illustrating just bioinformatics users personas (for Competency mapping into more roles/personas, please see the reference attached):
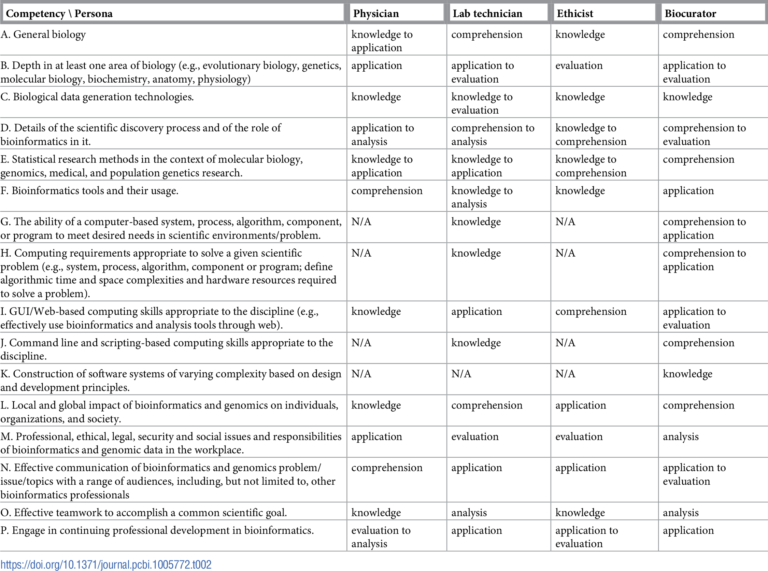 Competency mapping for bioinformatics users(Click to expand)
Competency mapping for bioinformatics users(Click to expand)
Please note: this competency framework uses original Bloom’s taxonomy, not a revised version we use in this course; however, the first three, lower cognitive levels are the same for both versions
The diagram below depicts the process of using a CF to design a bioinformatics course. While the comprehensive set of competencies as a whole is more applicable to longer courses and programmes, only a subset of competencies will normally be relevant for subject specific short courses.
 Diagram based on the Guidelines for developing and updating short courses and course programmes using the ISCB competency framework, with permission from Dr Nicky Mulder
Diagram based on the Guidelines for developing and updating short courses and course programmes using the ISCB competency framework, with permission from Dr Nicky Mulder
As depicted on the diagram, it is important to start with determining who your target audience is: what their previous knowledge is and whether their role matches approximately any of the roles/personas in the ISCB CF.
You can then find the appropriate role/persona in the corresponding tables (bioinformatics users, scientists or engineers) and check to which Bloom’s level this competency should be developed to. Please note that this CF offers one set of competencies and that your target audience might require some other professional competencies, related to other frameworks, so it is advisable to look at wider requirements and needs for your target audience.
With the list of competences and the levels you need to get your target audience to reach, you proceed with writing of the learning outcomes (Step 1.17) and defining the objectives/content to align with them. To be able to measure if your intended learning outcomes will be achieved, both formative and summative assessment need to be planned. Short course providers are likely to rely more on formative assessment, which will allow both learners and trainers to determine whether the required competency has been acquired.
The iterative nature of course design and development assumes that this process is never one off and never a strict algorithmic procedure. There should also exist different evaluation mechanisms that can be planned at the stage of course design, to help with deciding on changes and improvements.
Beyond competency frameworks
In recent years, the results of practical application of different competency frameworks had led some researchers and practitioners to suggest ways of improvement to bioinformatics competencies, so that it is possible to identify phases in competency development, and to provide guidance on the evidence required to assess whether some competency has been acquired.
One such example, Mastery Rubric for Bioinformatics (MR-Bi) by Rochelle E. Tractenberg et al. (2019) (reference attached below), is a somewhat wider framework that supports the development of specific KSAs, but also provides a structured trajectory for achieving bioinformatics competencies from the novice to expert level. It also emphasises the scientific method and the development of independent scientific reasoning and practice. It can be used to support complete course/programme or shorter training courses design, and to guide teaching and learning of specific KSA based on Principles of Andragogy (Step 1.9) of self-directed learning. For further reading, please find the reference attached in this Step.
Application of competency mapping in genomic medicine
Other competency frameworks have been developed, tailored to the specific needs of a particular profession or level. The National Health Services (NHS) Health Education England, UK competency mapping for genomics education used a competency framework for bioinformatics as it relates to genomic medicine. Using a training needs analysis, the competencies required by the clinical practitioners were identified https://www.genomicseducation.hee.nhs.uk/wp-content/uploads/2019/06/Clinical_Bioinformatics_Training.pdf
A similar competency-based approach was applied by the College of American Pathologists to develop a genomics curriculum for pathologists to be equipped with the competencies needed to practice successfully. Laudadio et al, 2014 firstly conducted a needs assessment survey to rate competency statements in 6 subject areas in pathology, namely Basic Genomics Concepts; Sample Acquisition; Testing and Interpretation; Patient Management and Reporting; Quality Assurance and Regulatory; and Ethical, Legal, and Social Issues. This enabled prioritisation of subject competency areas and refinement of the curriculum through identification of tasks that may be incorporated by pathologists in clinical practice. In addition, ideal active learning training strategies were also identified such as interactive formats and simulated cases. The competency map can also be useful for assessing individual learning needs and guide development of content and tools.
As an example the Basic Genomics Concepts subject area was elaborated as follows:
Competency statement – “Define targeted gene panel, whole genome sequencing, whole exome and whole transcriptome sequencing competency statement”
Sub-topics – Sequencing technologies; Targeted gene panels; Whole genome, exome, transcriptome sequencing
Expected tasks which demonstrate proficiency in the subject area:
- Compare and contrast Sanger and Next Generation Sequencing
- Describe the role of microarrays in the study of genomics (comparative genomic hybridization, RNA expression analysis, exome capture).
For each approach:
- Identify what is being sequenced and describe how that is being accomplished
- Describe pros and cons
- Explain to a clinical colleague the clinical significance of test results
Nembaware et al (2020) describe how the African Genomic Medicine Training Initiative (AGMT) developed a competency-based curriculum for genomic medicine in Africa. Starting with a general needs assessment, a targeted approach was applied to map competencies for nurses. Personas were developed based on common nurse specialisations in Africa and their roles and duties. This included a general nurse, a research nurse, community health nurse and a midwife. This was useful in developing appropriate teaching strategies and prioritising relevant content for developing a genomic medicine course. Topics included, genetic counselling, community engagement, genetics and genomics in patient care, addressing stigmas and misconceptions of genetics and genomics.
Share this
Train the Trainer: Design Genomics and Bioinformatics Training

Train the Trainer: Design Genomics and Bioinformatics Training


Reach your personal and professional goals
Unlock access to hundreds of expert online courses and degrees from top universities and educators to gain accredited qualifications and professional CV-building certificates.
Join over 18 million learners to launch, switch or build upon your career, all at your own pace, across a wide range of topic areas.
Register to receive updates
-
Create an account to receive our newsletter, course recommendations and promotions.
Register for free







