Home / Healthcare & Medicine / Bioinformatics / Making sense of genomic data: COVID-19 web-based bioinformatics / Assembly of SARS-CoV-2 genome and sequence alignment
This article is from the free online
Making sense of genomic data: COVID-19 web-based bioinformatics


Reach your personal and professional goals
Unlock access to hundreds of expert online courses and degrees from top universities and educators to gain accredited qualifications and professional CV-building certificates.
Join over 18 million learners to launch, switch or build upon your career, all at your own pace, across a wide range of topic areas.

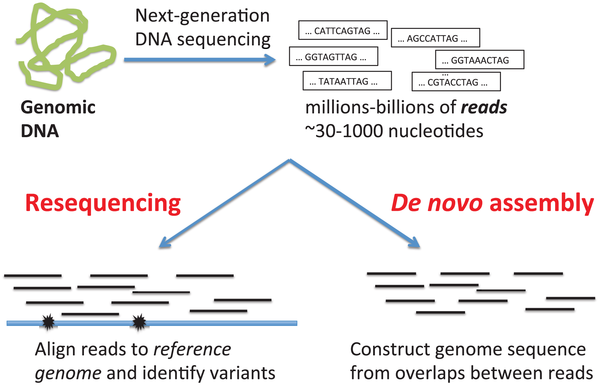
 C. Details in the main text”>
C. Details in the main text”>






