Checking the quality of amplicons
Genome sequencing of SARS-CoV-2 involves performing PCR using ARTIC or Midnight protocol. PCR amplification is the common step involved in both protocols. In order to confirm the successful PCR amplification, PCR amplicons need to be analysed. Before pooling the PCR products obtained from Pool A and B, the PCR amplicons are either loaded on agarose gel or QIAxcel to confirm successful amplification. The PCR products are then pooled and quantified using Qubit. You can review the Qubit method in step 1.21 Assessing the quality of a sample.
Depending on the information desired, there are different methods to analyse the products of a PCR reaction. Agarose gel electrophoresis, capillary gel electrophoresis and fragment analysis are the methods used to analyse the PCR amplicons. Mutation detection methods, such as denaturing gradient gel electrophoresis (DGGE) and temporal temperature gel electrophoresis (TTGE), which use acrylamide gel to assist with identifying mutations in the PCR product, are also available.
Quality checks of the SARS-CoV-2 amplicons can be performed by:
- Running the PCR amplicons – Slab gel electrophoresis, Capillary gel electrophoresis
- Quantifying of PCR amplicons – Qubit
Agarose gel Electrophoresis (Slab gel electrophoresis)
Agarose gel electrophoresis is a common technique to detect the presence or absence of the target sequence and the length of the fragment. Nucleic acid fragments are separated by their length while moving through an agarose matrix. By adding a dye or an intercalating agent like ethidium bromide (EtBr), these fragments can be visualised under ultraviolet light. The intensity of the band can be used to estimate the amount of product of a given molecular weight relative to a ladder. Gel electrophoresis also shows the specificity of the reaction, where the presence of multiple bands indicates secondary amplification products.
Slab gel electrophoresis (SGE) is a widely used technique for the analysis of PCR fragments. While SGE is a relatively inexpensive and easy-to-use technique, the amount of information, which can be derived with little effort from slab gels, is limited. Typically, size and concentration information is estimated by the scientist through visual comparison to the appropriate size and mass ladders which have been run in separate lanes on the gel. These estimations might be appropriate for experiments where a simple yes-or-no answer is adequate.
While gel electrophoresis is relatively easy to adopt in any laboratory, it has a number of disadvantages, including highly labour-intensive preparation of slab gels, and user exposure to hazardous chemicals such as ethidium bromide as a staining agent. Moreover, separation times can be rather long, depending on the experimental settings, and slab gels usually do not provide the high resolution required to meet today’s research demands.
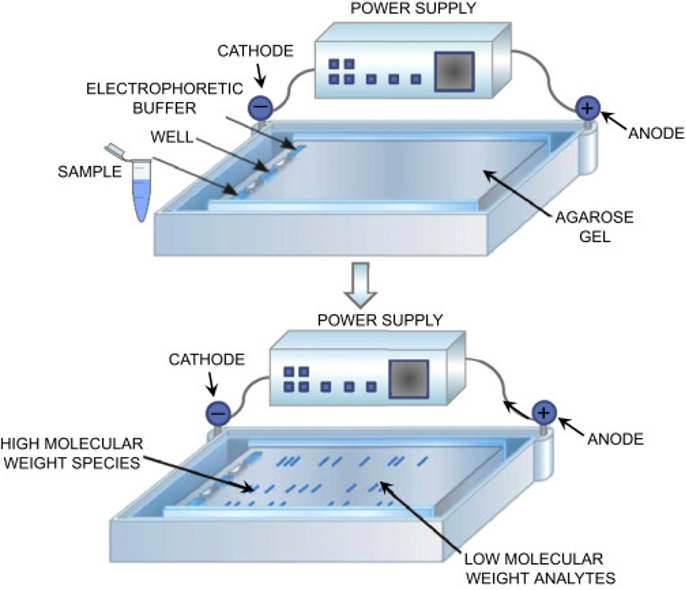
Figure 1 – Principle of Agarose gel electrophoresis. A tray containing the gel is positioned horizontally into a tank containing a buffer solution. A power supply is connected to the tank. Samples and molecular weights (aka, ladder) are loaded onto the cathode extremity of the gel. When the power supply is on, the negatively charged nucleic acid molecules are impulsed to migrate from cathode (-) to anode (+) through the gel. Low molecular weight analytes reach the anode extremity “faster” than the high molecular weight ones.
Capillary gel Electrophoresis (CGE)
Separation of analyte ions via differential migration in an electric field, coupled with the electro-osmotic flow of the mobile phase. Capillary electrophoresis (CE) is a process used to separate ionic fragments by size.
There are two different forms of capillary gel electrophoresis:
- Sequencer-based CGE
- QIAxcel-based CGE
Sequencer-based CGE (SCGE)
Sequencer-based capillary gel electrophoresis (SCGE) as an alternative to conventional gel electrophoresis, using 5′-end fluorescein-labelled primers and the AB 310 Genetic Analyzer (Applied Biosystems, Carlsbad, USA) with a 41-cm capillary loaded with a POP4 gel. This method was highly reproducible (the standard deviation of peak sizes was ±0.5 bp), independent of the reagents used, and had much higher discriminatory power than conventional agarose gel electrophoresis.
The use of fluorescein-labelled primers increases the sensitivity and, also, the cost of the method. SCGE requires access to a DNA sequencer, which may cause a delay if PCR products are referred to an external sequencing facility, and the cost of consumables per isolate is greater ($6 to $7, including primers, labels, PCR reagents, and SCGE, including a component for equipment cost) than for QCGE. SCGE requires the use of a more expensive instrument.
SCGE is not used for analysing the SARS-CoV-2 PCR amplicons, as SCGE requires fluorescent tagging of the primers.
QIAxcel-based CGE
The automated commercial CGE method, QIAxcel (Qiagen, Hilden, Germany), is used in many SARS-CoV-2 studies for analysing the PCR amplicons, which does not require the use of fluorescein-labelled primers.
QIAxcel is an automated electrophoresis platform that can deal with up to 96 samples per run with high efficiency and a turnaround time of 0.2 to 1.5h depending on the number of specimens. It is simple to perform, and potentially suitable for a clinical laboratory.
The results can be exported in either electropherogram or gel-view format. The major costs are for the setup of the QIAxcel system hardware and BioCalculator analysis software and for consumables (cartridges), making the cost per sample around $3 to $4. However, compared with SCGE, QCGE has limited sensitivity and discriminatory power, making it unable to clearly distinguish between amplicons with a 3-5bp difference.

Figure 2 – Sample separation using the QIAxcel Advanced System

Figure 3 – QIAxcel gel image of PCR amplicons with different sizes
Advantages and disadvantages of PCR amplicon analysis methods
| Method | Advantages | Disadvantages |
|---|---|---|
| Agarose gel electrophoresis | Inexpensive; Easy to use technique; Cost effective | Unsafe when EtBr is used; Low resolution; Time consuming; Labour intensive; Requires high concentration of Nucleic acids for detection |
| Capillary gel electrophoresis | Rapid analysis of up to 96 samples without manual intervention; Safe and Convenience with ready-to-use gel cartridges l; Robust results for nucleic acid concentrations as low as 0.1 ng/µl; Standardised and accurate analysis with a resolution down to 3–5 bp | Expensive compared to SGE |
Share this
A Practical Guide for SARS-CoV-2 Whole Genome Sequencing

A Practical Guide for SARS-CoV-2 Whole Genome Sequencing


Reach your personal and professional goals
Unlock access to hundreds of expert online courses and degrees from top universities and educators to gain accredited qualifications and professional CV-building certificates.
Join over 18 million learners to launch, switch or build upon your career, all at your own pace, across a wide range of topic areas.
Register to receive updates
-
Create an account to receive our newsletter, course recommendations and promotions.
Register for free







